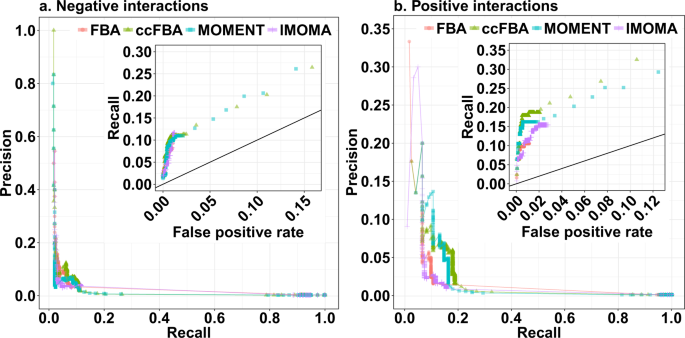
Flux balance analysis with or without molecular crowding fails to predict two thirds of experimentally observed epistasis in yeast | Scientific Reports

Improving the flux distributions simulated with genome-scale metabolic models of Saccharomyces cerevisiae - ScienceDirect

Metabolite Labeling prediction. The combination of 13C labeling data... | Download Scientific Diagram

Emergent sub-population behavior uncovered with a community dynamic metabolic model of Escherichia coli diauxic growth | bioRxiv

Geometric illustration of the eMOMA method. Red-colored area represents... | Download Scientific Diagram

Improving the flux distributions simulated with genome-scale metabolic models of Saccharomyces cerevisiae - ScienceDirect

A variant of flux balance analysis shows consistency with proteomic and... | Download Scientific Diagram

Assessing Escherichia coli metabolism models and simulation approaches in phenotype predictions: Validation against experimental data - Costa - 2018 - Biotechnology Progress - Wiley Online Library
Emergent sub-population behavior uncovered with a community dynamic metabolic model of Escherichia coli diauxic growth Supplemen
PLOS Computational Biology: Stoichiometric Representation of Gene–Protein–Reaction Associations Leverages Constraint-Based Analysis from Reaction to Gene-Level Phenotype Prediction
PLOS Computational Biology: Stoichiometric Representation of Gene–Protein–Reaction Associations Leverages Constraint-Based Analysis from Reaction to Gene-Level Phenotype Prediction

The relationship between the target chemical production flux and the... | Download Scientific Diagram
PLOS Computational Biology: Stoichiometric Representation of Gene–Protein–Reaction Associations Leverages Constraint-Based Analysis from Reaction to Gene-Level Phenotype Prediction

Assessing Escherichia coli metabolism models and simulation approaches in phenotype predictions: Validation against experimental data - Costa - 2018 - Biotechnology Progress - Wiley Online Library

FBA classes are consistent with omic data. Simulations for each growth... | Download Scientific Diagram

In silico identification of metabolic engineering strategies for improved lipid production in Yarrowia lipolytica by genome-scale metabolic modeling | Biotechnology for Biofuels | Full Text

Improving the flux distributions simulated with genome-scale metabolic models of Saccharomyces cerevisiae – topic of research paper in Biological sciences. Download scholarly article PDF and read for free on CyberLeninka open science
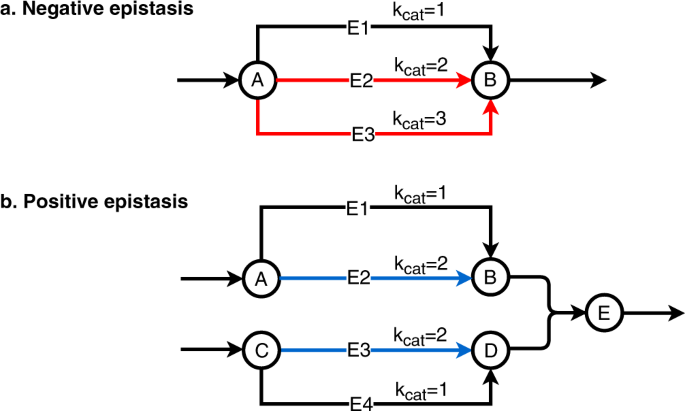
Flux balance analysis with or without molecular crowding fails to predict two thirds of experimentally observed epistasis in yeast | Scientific Reports

Grid-based computational methods for the design of constraint-based parsimonious chemical reaction networks to simulate metabolite production: GridProd | SpringerLink

![Full text] Designing metabolic engineering strategies with genome-scale metabolic | AGG Full text] Designing metabolic engineering strategies with genome-scale metabolic | AGG](https://www.dovepress.com/cr_data/article_fulltext/s58000/58494/img/fig1.jpg)
![Full text] Designing metabolic engineering strategies with genome-scale metabolic | AGG Full text] Designing metabolic engineering strategies with genome-scale metabolic | AGG](https://www.dovepress.com/cr_data/article_fulltext/s58000/58494/img/Table1.jpg)
